First time users of the DNC should meet with Drs. Neil or Shimony, for an initial consultation and discussion of the user’s needs. Depending on the project, the users will be referred to the appropriate unit leaders for further discussions relevant to the expertise and deliverables of that unit and the formulation of a research plan. Each unit will provide, as appropriate: consultation regarding research protocol development; technical assistance in carrying out experiments; resources for data analysis; support for statistical analyses; training on MRI scanner operation or a member of the unit to run the scanner; assistance with grant applications and manuscript preparation.
| UNIT | SERVICE | COST / UNIT |
| HIU | Initial Consultation for Human Imaging Study | Free Initial 2- Hour Consultation |
| AIU | Initial Consultation for Rodent Imaging Study | Free Initial 2-Hour Consultation |
| General | Secondary Analysis of Major Datasets: ABCD | Custom |
| General | Secondary Analysis of Major Datasets: eLAB | Custom |
| General | Secondary Analysis of Major Datasets: IBIS | Custom |
| HIU | Analysis of New Imaging Data: Human | 60 / Hour |
| AIU | Analysis of New Imaging Data: Rodent | 60 / Hour |
| NU | Neuroinformatics / Data Storage-Management: Human | Custom |
| NU | Neuroinformatics / Data Storage-Management: Rodent | Custom |
| HIU | Research Transformation of Clinical Scans: Human | Custom |
| AIU | Animal handling, surgery and other procedures | 60 / Hour |
| AIU | Technical Assistance, rodent imaging | No Cost |
| HIU | Technical assistance, human developmental brain imaging | No Cost |
| HIU | Mock Scanning | No Cost |
| AIU | Rodent Brain Imaging | $105 / Scanner Hour |
| HIU | Technical Assistance, Respiract device setup | No Cost |
| HIU | Assist in scanning, Respiract device | Custom |
For more information about the services of the Developmental Neuroimaging Core, please contact:
Joshua Shimony: shimonyj@wustl.edu; 314-362-5950
Jeffrey Neil: neil@wustl.edu; 314-454-6120
Animal Imaging Unit
The AIU provides assistance with animal specific imaging methods, including anesthesia, restraint, scanner operation, and surgical techniques.
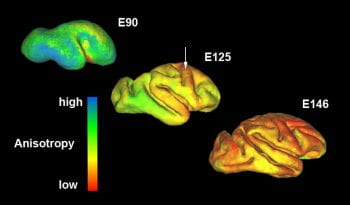
Imaging sequences that are currently supported by the AIU include
- standard anatomical imaging (e.g., T1, T2, and T2*)
- the following advanced MR techniques: Diffusion Weighted (DWI) and Diffusion Tensor Imaging (DTI) with tractography
- Perfusion imaging, including Dynamic Susceptibility Contrast (DSC), Dynamic Contrast Enhancement (DCE), and Arterial Spin Labeling (ASL)
- Magnetization Transfer (MT)
- resting state functional connectivity MRI (rs-fcMRI)
- Gradient Echo Plural Contrast Imaging (GEPCI)
- techniques for measuring tissue PO2
- MR spectroscopy
The Biomedical MR Lab (BMRL) was recently awarded an NIH-funded S10 high-end instrumentation grant to purchase a state-of-the-art Bruker BioSpec 94/20 USR small animal imaging system, which will replace one of its current 4.7-T systems. We anticipate it will be installed and available to support IDDRC research by summer 2020. The Bruker scanner operates at a field strength of 9.4 T and has a 200-mm clear bore. A number of RF coils will be available. Included among them will be a 4-element CryoProbe receiver for adult mouse head that will increase signal-to-noise ratio by a factor of 2-4x and be useful for a number of studies, including fMRI.
Human Imaging Unit
Imaging research requires a high level of specialized expertise that continues to evolve with advancements in the field. This expertise is not typically available to clinical investigators and will be made available to IDDRC investigators through the HIU.
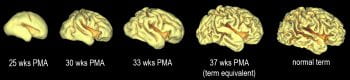
The MR imaging modalities supported include, but are not limited to: conventional structural imaging, including post-acquisition segmentation and volumetry, DTI and tractography, Task-based fMRI and rs-fcMRI, and MR Perfusion imaging using various techniques. Analysis of neuroimaging data is complex and dependent upon the particular imaging modality employed. The unit will provide services related to multiple imaging data formats, including some that have been developed locally (i.e., “4dfp”). The HIU also has expertise using versatile imaging software analysis packages such as FSL (Oxford University, Oxford, UK) and FreeSurfer (Harvard University, Cambridge, MA). The HIU provides a full range of image analysis services for the imaging modalities listed above.
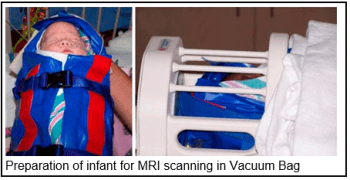
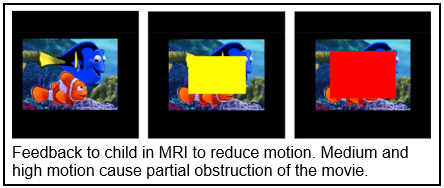
Services to improve the success rates for infant and early childhood imaging have been expanded substantially. Recent studies have demonstrated that with careful home preparation, proper sound attenuation, and attention to the child’s sleep cycle, successful production of high quality imaging data is possible at any age. Dr. Botteron and her team can provide their valuable expertise in scanning unsedated infants, toddlers, and young children to IDDRC investigators by consultation, the development of procedural and behavioral protocols for automatic implementation across all studies, and direct supervision for initial groups of subjects.
As needed, the unit will also develop additional novel protocols for scanning subjects with unique developmental disorders or older age groups than our current studies (e.g., low functioning autism, Wolfram syndrome, Down syndrome, or severe prematurity) who may require more intensive pre-scanning behavioral training that is tailored to their specific condition.’

Dr. Culver can provide a full range of services for human and rodent optical neuroimaging, from initial consultation to study implementation. Dr. Culver and his team will work directly with IDDRC investigators to acquire optical neuroimaging data. The Culver laboratory developed, built and currently operates six HD-DOT instruments, three fixed location systems with large channel counts (DOT-A has 96 sources and 96 detectors, DOT-B & DOT-C each have128 sources and 128 detectors). We also have three portable clinical instruments (DOT-D has 36 sources and 36 detectors, DOT-E has 32 sources and 48 detectors, and DOT-F has 72 sources and 72 detectors). Data acquisition services include adapting these existing HD-DOT systems to the user’s needs. DOT–C is specifically optimized for children aged 12 to 48 months, DOT–F is optimized for children from infancy through nine months. DOT–D is the most portable system and is currently being used in Cali, Columbia to collect data on children with malnutrition. Systems C, D and F are supported by funding from the Bill and Melinda Gates Foundation.
Neuroinformatics Unit
The Neuroinformatics Unit can provide
- the necessary data management, automated image processing/analysis, and computing infrastructure to allow IDDRC investigators to undertake developmental neuroimaging studies with minimal barriers to success,
- data packaging that facilitates access to developmental researchers, and
- integrate the imaging informatics into broader informatics systems as needed
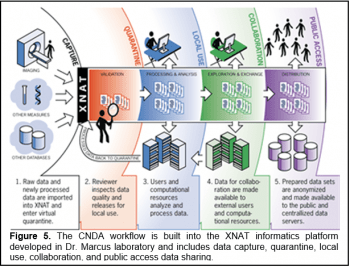
The Neuroinformatics Unit provides a secure data management system to capture, organize, annotate, process, and share neuroimaging and related data for IDDRC investigators through the CNDA platform. The CNDA is integrated with all research scanners and radiology information systems to enable streamlined and automated data archiving. The CNDA is supported by full-time helpdesk and technical support staff, online tutorials and other documentation, and regular live training sessions.
The data management services focus on five key functions: data capture, quality control, productivity, collaboration, and data sharing:
Data capture. Acquired data from IDDRC investigators will be automatically imported into study-specific projects. Once received by the CNDA, the headers of all incoming DICOM data will be parsed to extract the research protocol, subject, visit, acquisition details, and other metadata. The files will be placed in the CNDA’s secure permanent archive, and the metadata will be written to the CNDA’s searchable database. Additional tools, including a DICOM browser and a web-based uploader, are available for importing data that are not available at the scanner. Finally, we will support multi-center studies, where data are obtained in remote imaging facilities, by providing a web-based upload tool.
Quality Control. If desired by the investigator, data can be placed into a virtual quarantine until they are inspected and approved by an authorized user. If errors in a quarantined record are identified, they can be corrected using XNAT’s online forms or traced back to their point of origin. Data are released from quarantine by users with appropriate security privileges. Additional quality control tools include a DICOM validator that compares metadata fields included in uploaded images with a prescribed protocol and generates a validation report. Manual QC forms allow study members to enter image quality reviews.
Productivity. The CNDA includes a built-in image viewer, and the CNDA web services can be used to integrate XNAT-hosted data into external viewers such as 3D Slicer and Connectome Workbench. The CNDA also includes a search engine for identifying data sets of interest.
Collaboration. The IDDRC imaging informatics platform is fundamentally a collaboration environment for team science. The CNDA serves as a central hub around which collaborators exchange data and common methods. The basic organizational unit in the CNDA is the project. Investigators have complete control over which users can access their projects.
Data sharing. IDDRC investigators are increasingly interested in data sharing. The CNDA includes a data sharing pipeline that removes sensitive metadata, obscures facial features in the image content, converts images to NIFTI format, and generates BIDS-formatted data bundles . Once the data sharing pipeline has been executed, the data can be openly shared with the broader scientific community through public access data repositories such as XNAT Central and OpenNeuro.
The CNDA enables standard and IDDRC investigator modified pipelines to be executed on the Center’s computing cluster using Docker and Singularity containers, which provide a compact, portable and platform-independent mechanism for packaging the large battery of applications, tools, and scripts upon which the pipelines are based. Execution of containerized pipelines on the CNDA utilizes the XNAT Container Service, which automates and manages running of containers on the computing infrastructure. Several pipelines have been already been developed specifically for IDDRC users, such as that for processing of rs-fcMRI data. There is ongoing work to further develop, test, and deploy automated pipelines to process data for the IDDRC.
The Neuroinformatics Unit operates a scalable, high availability Linux-based computing infrastructure to support the Center’s data management systems, pipeline execution cluster, user workstations, and computing lab.
The Neuroinformatics Unit provides ongoing training and education opportunities to the IDDRC community through seminars hosted by the Department of Radiology, including a monthly program that includes internal and external speakers with a focus on both neuroimaging research and methodological advances. The seminar is regularly attended by over 75 faculty, trainees, and staff, including many from IDDRC-affiliated laboratories. The Neuroinformatics Unit will also sponsor formal training programs, including an annual “Introduction to MRI” program led by Dr. Shimony and short courses focused on particular methods and applications. Recent short courses have included Introduction to FSL, Introduction to R, and Introduction to High Performance Computing.